
Loading and checking data
Johannes Krietsch
Source:vignettes/load_and_check_data.Rmd
load_and_check_data.RmdGood to know
tools4watlas is based on data.table to be
fast and efficient. A key feature of data.table is
modification in place, where data is changed without making a copy. To
prevent this (whenever it is not desired) use the function
copy() to make a true copy of the data set. Basic knowledge
about data.table
is helpful, but not necessary, when working with
tools4watlas.
Getting data
WATLAS data can either be loaded from a local SQLite database or a remote SQL database server. To do so, first select the tags and time period for which to extract data.
Use the tags_watlas_all.xlsx file (including metadata of
all tags) or for collaborators the tags_watlas_subset.xlsx
(including a subset of metadata) to select the desired tags. Here shown
with the example data in tools4watlas.
Loading the tags_watlas_subset.xlsx will provide a table
with the following columns:
| Column | Description |
|---|---|
| year | Year in which the bird was caught |
| species | Species common name |
| tag | Tag ID with 4 digits |
| rings | Metal ring number |
| crc | Colour ring combo |
| release_ts | Release time stamp in CET |
| catch_location | Location where the bird was caught |
Select the desired tags and time period
# file path to the metadata
fp <- system.file(
"extdata", "tags_watlas_subset.xlsx", package = "tools4watlas"
)
# load meta data
all_tags <- readxl::read_excel(fp, sheet = "tags_watlas_all") |>
data.table()
# subset desired tags using data.table
# (for example all tags from 2023)
tags <- all_tags[year == 2023]$tag
# time period for which data should be extracted form the database (in CET)
from <- "2023-09-21 00:00:00"
to <- "2023-09-25 00:00:00"Extract data from local SQLite file
First, the path and file name of the local SQLite database need to be provided. Then, with the established connection, the database can be queried for the selected tags and period. Here we will load the tagging data in one data.table.
# database connection
sqlite_db <- system.file(
"extdata", "watlas_example.SQLite", package = "tools4watlas"
)
con <- RSQLite::dbConnect(RSQLite::SQLite(), sqlite_db)
# load data from database
data <- atl_get_data(
tags,
tracking_time_start = from,
tracking_time_end = to,
timezone = "CET",
use_connection = con
)
# close connection
RSQLite::dbDisconnect(con)Alternatively, extract from remote SQL-database
To safely work with database credentials one option is to store them as environmental variables in R. This allows for example to have scripts on GitHub without sharing them. Restart R after adding variables to be able to access them. Ask Allert for the host, username and password and then add them in your environment like this:
# open .Renviron to edit
file.edit("~/.Renviron")
# add variables
host = "host"
username = "username"
password = "password"
# access variables (example)
Sys.getenv("variable_name")atl_get_data does the same as when connecting to a local
SQLite database when connecting to a remote database. In this example
(chunk not run and only shown) we load the last three days of data from
all tags of 2024. Host, username and password are specified as
environmental variables in this example, but can also be specified
directly.
Since we want the up to date metadata we load the
tags_watlas_all.xlsx from the “WATLAS” SharePoint folder:
Documents/data/. Either specify the path to your local copy
of this folder or add the path for your user in the
atl_file_path() function.
# file path to WATLAS teams data folder
fp <- atl_file_path("watlas_teams")
# load meta data
all_tags <- readxl::read_excel(
paste0(fp, "tags/tags_watlas_all.xlsx"),
sheet = "tags_watlas_all"
) |>
data.table()
# subset all tags from 2024
tags <- all_tags[year == 2024]$tag
# select N last days to get data from
days <- 3
from <- (Sys.time() - 86400 * days) |> as.character()
to <- (Sys.time() + 3600) |> as.character()
# load data from database
data <- atl_get_data(
tags,
tracking_time_start = from,
tracking_time_end = to,
timezone = "CET",
host = Sys.getenv("host"),
database = "atlas2024",
username = Sys.getenv("username"),
password = Sys.getenv("password")
)Data explanation
The resulting loaded WATLAS data will be a data.table
with the following columns:
| posID | tag | time | datetime | x | y | nbs | varx | vary | covxy |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 3027 | 1695438802 | 2023-09-23 03:13:22 | 650692.8 | 5902549 | 3 | 46.97 | 392.42 | 126.62 |
| 2 | 3027 | 1695438805 | 2023-09-23 03:13:25 | 650705.6 | 5902576 | 3 | 49.09 | 460.84 | 141.21 |
| 3 | 3027 | 1695438808 | 2023-09-23 03:13:28 | 650691.6 | 5902536 | 3 | 58.18 | 471.81 | 155.89 |
| 4 | 3027 | 1695439189 | 2023-09-23 03:19:49 | 650728.6 | 5902571 | 3 | 49.97 | 441.46 | 138.20 |
| 5 | 3027 | 1695439192 | 2023-09-23 03:19:52 | 650721.0 | 5902556 | 3 | 5.34 | 28.16 | 8.58 |
| 6 | 3027 | 1695439195 | 2023-09-23 03:19:55 | 650721.1 | 5902559 | 3 | 5.55 | 35.03 | 10.43 |
| Column | Description |
|---|---|
| posID | Unique number for localizations |
| tag | 4 digit tag number (character), i.e. last 4 digits of the full tag number |
| time | UNIX time (seconds) |
| datetime | Datetime in POSIXct (UTC) |
| x | X-cordinates in meters (UTM 31 N) |
| y | Y-cordinates in meters (UTM 31 N) |
| nbs | Number of Base Stations used in calculating coordinates |
| varx | Variance in estimating X-coordinates |
| vary | Variance in estimating Y-coordinates |
| covxy | Co-variance between X- and Y-coordinates |
Remove data before release
# correct time zone to CET and change to UTC
all_tags[, release_ts := force_tz(as_datetime(release_ts), tzone = "CET")]
all_tags[, release_ts := with_tz(release_ts, tzone = "UTC")]
# join release_ts with data
all_tags[, tag := as.character(tag)]
data[all_tags, on = "tag", `:=`(release_ts = i.release_ts)]
# exclude positions before the release
data <- data[datetime > release_ts]
# remove release_ts column
data[, release_ts := NULL]Add species column (or other relevant columns)
If we are working with multiple species, then we can join the species from the metadata. In this case I want the species to be the first row of the table.
# join with species data
all_tags[, tag := as.character(tag)]
data[all_tags, on = "tag", `:=`(species = i.species)]
# make species first column
setcolorder(data, c("species", setdiff(names(data), c("species"))))
# order data.table
setorder(data, species, tag, time)We can also add other metadata by merging whatever column we want (e.g. color rings and catch location). However, when working with large data sets, it is advised to only add columns that are necessary. This can also always be done whenever needed. This is just an example and Here we delete the columns again.
# join with metal rings, color rings and catch location
all_tags[, tag := as.character(tag)]
data[all_tags, on = "tag", `:=`(
rings = i.rings,
crc = i.crc,
catch_location = i.catch_location
)]
# delete columns
data[, c("rings", "crc", "catch_location") := NULL]Save data
At this point it might be good to save the raw data, as extrating the
data from the database can take a long time with big datasets. A
convenient and fast way is to use fwrite from the
data.table package. By including yaml = TRUE
we make sure the data stays in the same format when we load it again.
Change the file path when running this example.
# save data
fwrite(data, file = "../inst/extdata/watlas_data_raw.csv", yaml = TRUE)Check data
Data summary
Here we simply check for how many individuals we have data and how many positions by tag and date we have.
# load data
data <- fread("../inst/extdata/watlas_data_raw.csv", yaml = TRUE)
# data summary
data_summary <- atl_summary(data, id_columns = c("species", "tag"))
# N individuals with tagging data
data_summary |> nrow()## [1] 8
# N by species
data_summary[, .N, by = species]## species N
## <char> <int>
## 1: bar-tailed godwit 1
## 2: curlew 1
## 3: dunlin 1
## 4: oystercatcher 1
## 5: red knot 1
## 6: redshank 1
## 7: sanderling 1
## 8: turnstone 1
# show head of the table
data_summary |> knitr::kable(digits = 2)| species | tag | n_positions | first_data | last_data | days_data | min_gap | max_gap | max_gap_factor | fix_rate |
|---|---|---|---|---|---|---|---|---|---|
| bar-tailed godwit | 3063 | 12614 | 2023-09-23 03:27:49 | 2023-09-23 22:24:55 | 0.8 | 3 | 2541 | 42.4 min | 0.55 |
| curlew | 3100 | 8423 | 2023-09-23 04:21:46 | 2023-09-23 21:41:16 | 0.7 | 3 | 16145 | 4.5 hours | 0.41 |
| dunlin | 3212 | 3856 | 2023-09-23 00:00:00 | 2023-09-23 23:59:56 | 1.0 | 8 | 4784 | 1.3 hours | 0.36 |
| oystercatcher | 3158 | 12533 | 2023-09-23 00:00:01 | 2023-09-23 23:59:57 | 1.0 | 3 | 3060 | 51 min | 0.44 |
| red knot | 3038 | 15959 | 2023-09-23 00:00:01 | 2023-09-23 23:59:57 | 1.0 | 3 | 2850 | 47.5 min | 0.55 |
| redshank | 3027 | 15859 | 2023-09-23 03:13:22 | 2023-09-23 22:24:26 | 0.8 | 3 | 2523 | 42 min | 0.69 |
| sanderling | 3288 | 8131 | 2023-09-23 00:00:03 | 2023-09-23 23:59:54 | 1.0 | 6 | 2622 | 43.7 min | 0.56 |
| turnstone | 3188 | 10195 | 2023-09-23 00:00:45 | 2023-09-23 23:41:50 | 1.0 | 3 | 5742 | 1.6 hours | 0.36 |
| Column | Description |
|---|---|
| species | Species |
| tag | Tag number |
| n_positions | Number of positions |
| first_data | Datetime first localization (UTC) |
| last_data | Datetime last localization (UTC) |
| days_data | Days of data |
| min_gap | Minimum time interval between localizations (interval of the tag in seconds) |
| max_gap | Maximum time interval between localizations (largest gap in seconds) |
| max_gap_factor | Maximum time interval between localizations as factor (in seconds, minutes, hours, or days) |
| fix_rate | Fix rate (=1 if every min_gap has a localization
between first_data and last_data) |
Plot the number of positions by day.
# add date
data[, date := as.Date(datetime)] |> invisible()
# N positions by species and day
data_subset <- data[, .N, by = .(tag, date)]
# plot data
ggplot(data_subset, aes(x = date, y = tag, fill = N)) +
geom_tile() +
scale_fill_viridis(
option = "A", discrete = FALSE, trans = "log10", name = "N positions",
breaks = trans_breaks("log10", function(x) 10^x),
labels = trans_format("log10", math_format(10^.x)),
direction = -1
) +
labs(x = "Date", y = "Tag") +
theme_classic()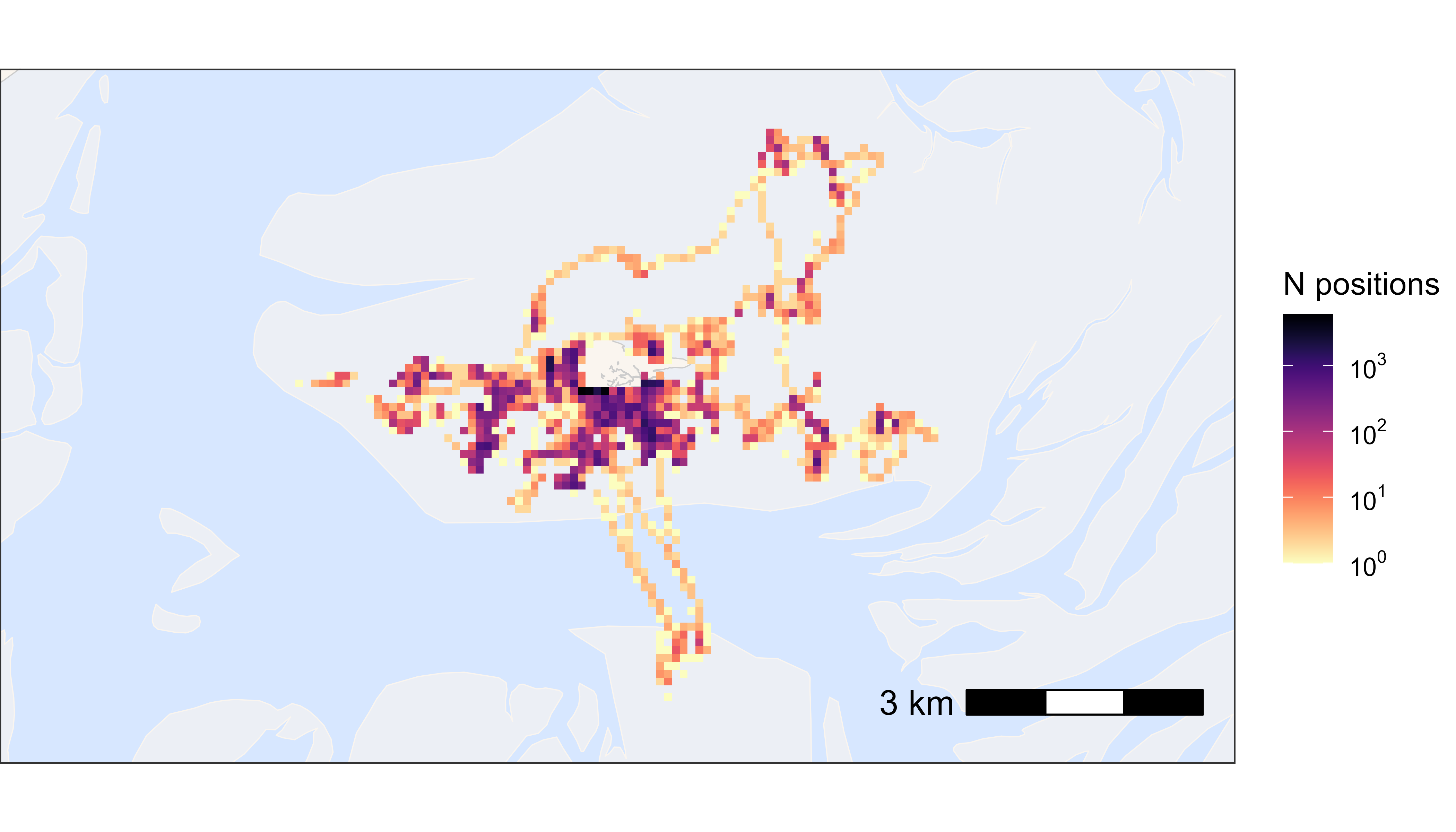
Number of positions per day by tag
Plot overview of the data
Now we want to see the data on a map. With large datasets it is convenient to plot heatmaps, as plotting for example 40 million points would result in a lot of over plotting and makes the plotting slow. If the dataset is small, one can obviously also plot the data by tag number or in other ways (see vignette plotting data).
# create basemap
bm <- atl_create_bm(data, buffer = 800)
# round data to 1 ha (100x100 meter) grid cells
data[, c("x_round", "y_round") := list(
plyr::round_any(x, 100),
plyr::round_any(y, 100)
)]
# N by location
data_subset <- data[, .N, by = c("x_round", "y_round")]
# plot heatmap
bm +
geom_tile(
data = data_subset, aes(x_round, y_round, fill = N),
linewidth = 0.1, show.legend = TRUE
) +
scale_fill_viridis(
option = "A", discrete = FALSE, trans = "log10", name = "N positions",
breaks = trans_breaks("log10", function(x) 10^x),
labels = trans_format("log10", math_format(10^.x)),
direction = -1
)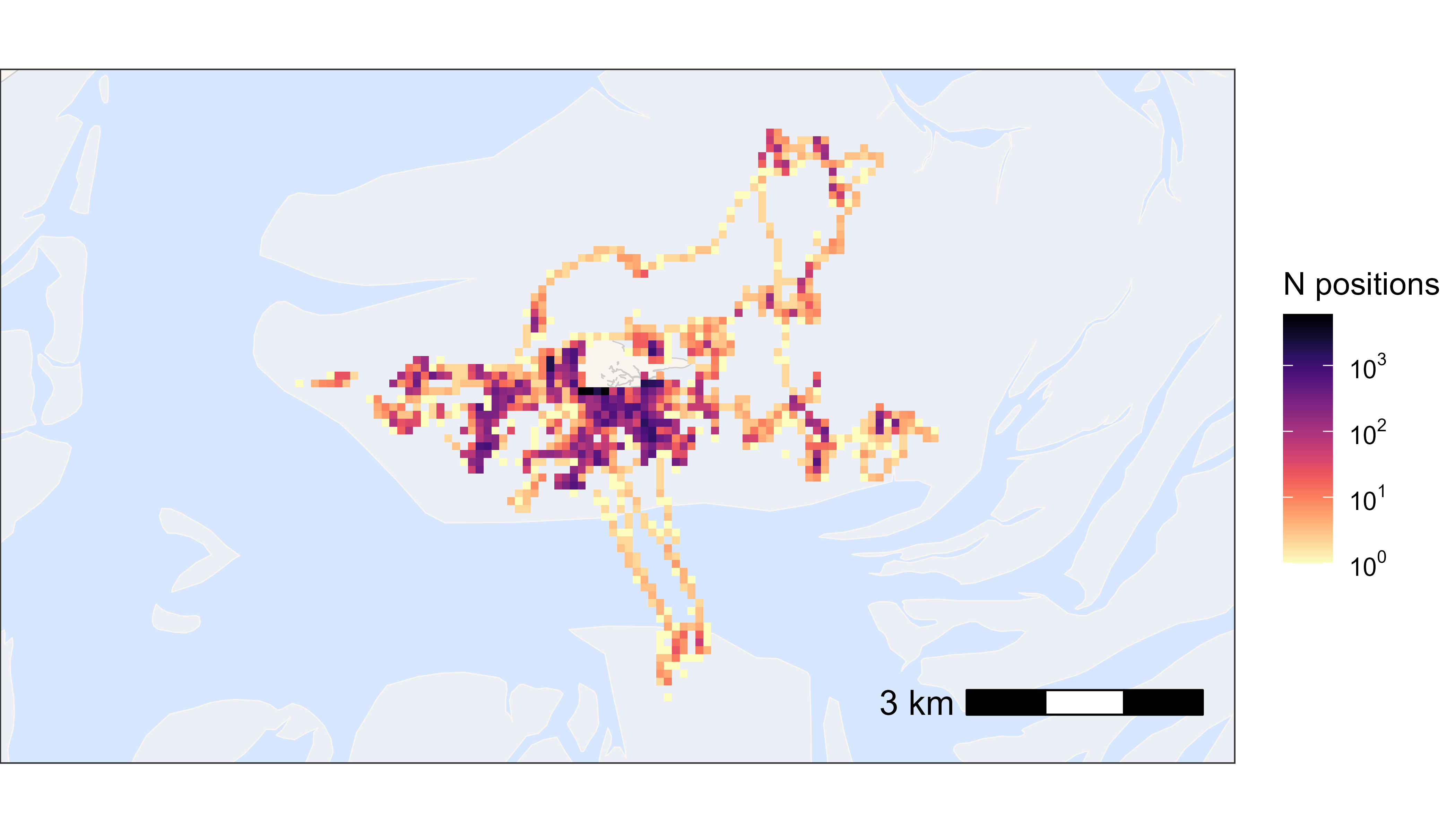
Heatmap of all positions